Exercises part III: statistical inference#
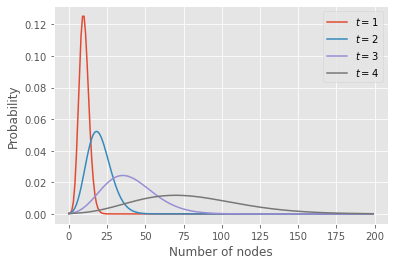
The code below extract the probability distribution for the number of nodes at a few time points for a branching process with a Poisson offspring distribution.
import numpy as np
import matplotlib.pyplot as plt
plt.style.use(['ggplot'])
R = 2.
Q = lambda x: np.exp(R*(x-1))
G0 = lambda x: x**5
N = 200
n = np.arange(N)
c = np.exp(2*np.pi*1j*n/N)
tset = {1,2,3,4}
x = c.copy()
for t in range(max(tset)+1):
if t in tset:
pn = abs(np.fft.fft(G0(x))/N)
plt.plot(n,pn, label=fr"$t = {t}$")
x = Q(x)
plt.legend()
plt.ylabel('Probability')
plt.xlabel('Number of nodes')
plt.show()
Tip
Remember to adapt the support to avoid aliasing effects.
Posterior distribution on the reproduction number#
Let us assume that we have an emerging outbreak. To simplify the situation, let us assume that the generation time is always a week, and infectious individuals generate a number of secondary case according to a Poisson distribution with PGF of the form
We can model this situation using a branching process, and we want to infer \(R\), the reproduction number.
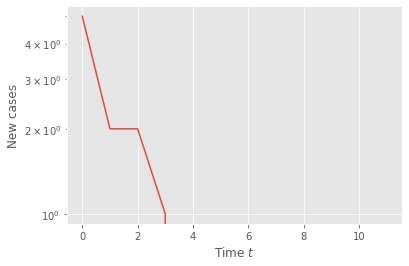
Run the code below to get a time series—this will represent the data available.
from secret_code import *
T = 12 #length of the time series
t = np.arange(T)
n = poisson_branching_process(T)
plt.semilogy(t,n)
plt.xlabel(r"Time $t$")
plt.ylabel(r"New cases")
plt.show()
Reusing the code above for the PGF, estimate the posterior distribution on \(R\).
Tip
As a first step, use only the first and last data points, \(n_0 = 1\) and \(n_T\). You have that \(G_0(x) = x^{n_0}\), and you can calculate \(G_T(x)\) (and the associated probability distribution) from the recursion implemented in the code above.
The posterior distribution is \(P(R|n_0,n_T) \propto P(n_T| n_0, R) P(n_0,R)\). We could choose different prior distribution, but right now let us assume that \(P(n_0,R)\) is simply some constant \(C\).
Since the likelihood \(P(n_T| n_0, R)\) is generated by \(G_T(x)\), we can evaluate the posterior distribution for a few values of \(R\) on an interval and plot the resulting curve.
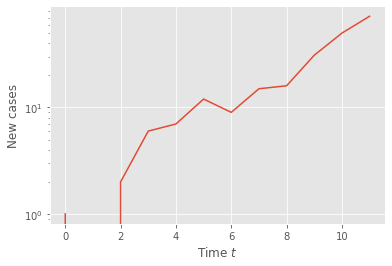
To make it a bit more realistic, apply your code to the following time series, where only a certain fraction of the cases are detected.
from secret_code import *
T = 12 #length of the time series
t = np.arange(T)
n = poisson_branching_process_with_noise(T)
plt.semilogy(t,n)
plt.xlabel(r"Time $t$")
plt.ylabel(r"New cases")
plt.show()
Try using more data points and breaking down the likelihood in a product (see below). Is the inference better?
Note
In the limit \(\tau = 1\), one could argue (successfully) that PGFs are not necessary here. Indeed, \(P(n_{t}|n_{t-1},R)\) is a Poisson distribution by the additive property of Poisson random variables. While this is true in this very simple exercise, realistic situations will lead to a much more complex likelihood which either require PGFs, or an approximation scheme making use of simulation like approximate Bayesian computation (ABC).